BioMedLM
Authors: Elliot Bolton and David Hall and Michihiro Yasunaga and Tony Lee and Chris Manning and Percy Liang
We introduce a new 2.7B parameter language model trained on biomedical literature which delivers an improved state of the art for medical question answering. (Note: This model was previously known as PubMedGPT.)
Introduction
Stanford CRFM has recently been studying domain-specific foundation models, i.e. models that are trained exclusively, or nearly exclusively, on data from a particular subdomain such as medicine or law. We believe this investigation will allow us to better understand how data composition impacts foundation models and produce more accurate and efficient systems for domain-specific downstream tasks.
We partnered with MosaicML to build BioMedLM, a new language model trained exclusively on biomedical abstracts and papers. This GPT-style model can achieve strong results on a variety of biomedical NLP tasks, including a new state of the art performance of 50.3% accuracy on the MedQA biomedical question answering task.
Today we are excited to make this model available to the community. As an autoregressive language model, BioMedLM is also capable of natural language generation. However, we have only begun to explore the generation capabilities and limitations of this model, and we emphasize that this model’s generation capabilities are only for research purposes and not suitable for production. In releasing this model, we hope to advance both the development of biomedical NLP applications and best practices for responsibly training and utilizing domain-specific language models; issues of reliability, truthfulness, and explainability are top of mind for us. We hope lessons learned from training this biomedical model can be applied to other domains such as law or finance.
Model Architecture
BioMedLM is an autoregressive language model with 2.7B parameters. It uses the standard GPT-2 architecture with the following settings:
| Parameter | Value |
|---|---|
| hidden size | 2560 |
| heads | 20 |
| layers | 32 |
| vocab size | 28896 |
| sequence length | 1024 |
The model uses a custom tokenizer trained on the PubMed Abstracts. When building domain specific models we have found it important to use a tokenizer trained on in-domain text to maximize performance on downstream tasks. A key benefit is that common biomedical terms are represented as entire tokens.
For instance, all of these following terms are tokenized into single tokens by the biomedical tokenizer and multiple tokens by the standard GPT-2 tokenizer:
| Word | Tokenization |
|---|---|
| chromatography | chrom/atography |
| cytotoxicity | cyt/ot/oxicity |
| Immunohistochemistry | Imm/un/oh/ist/ochemistry |
| photosynthesis | photos/ynthesis |
| probiotic | prob/iotic |
This allows the model to encode information about these concepts in their individual token representations rather than spread out across subword tokens like “oh” shared with many other terms.
The model code used during training was modified to utilize Flash Attention (Dao et al. 2022). This was an important modification which nearly halves total training time, making training a model of this scale possible for our team.
Training
BioMedLM was trained on all the PubMed abstracts and full documents from The Pile.
The model was trained on MosaicML Cloud, a platform designed for large workloads like LLMs. Using the Composer training library and PyTorch FSDP, it was easy to enable multi-node training across 128 A100-40GB GPUs, and the total run was completed in ~6.25 days. For more details on how we engineered and orchestrated training, see MosaicML’s companion blog post.
The model was trained with batch size=1024 and sequence length=1024 for 300B tokens using Decoupled AdamW with the following settings:
| Parameter | Value |
|---|---|
| lr | 1.6e-4 |
| eps | 1e-8 |
| betas | [0.9, 0.95] |
| weight decay | 1.6e-5 |
The training process was very smooth and did not suffer from any divergences.
As we were preparing the training run, we were unsure of the benefits of training out to 300B tokens for language model perplexity and downstream task performance. While most models of this scale (e.g. GPT Neo 2.7B) are trained to 300-400B tokens, the datasets those models use are vastly larger than PubMed. For instance, The Pile is 8x the size of its PubMed subcorpora.
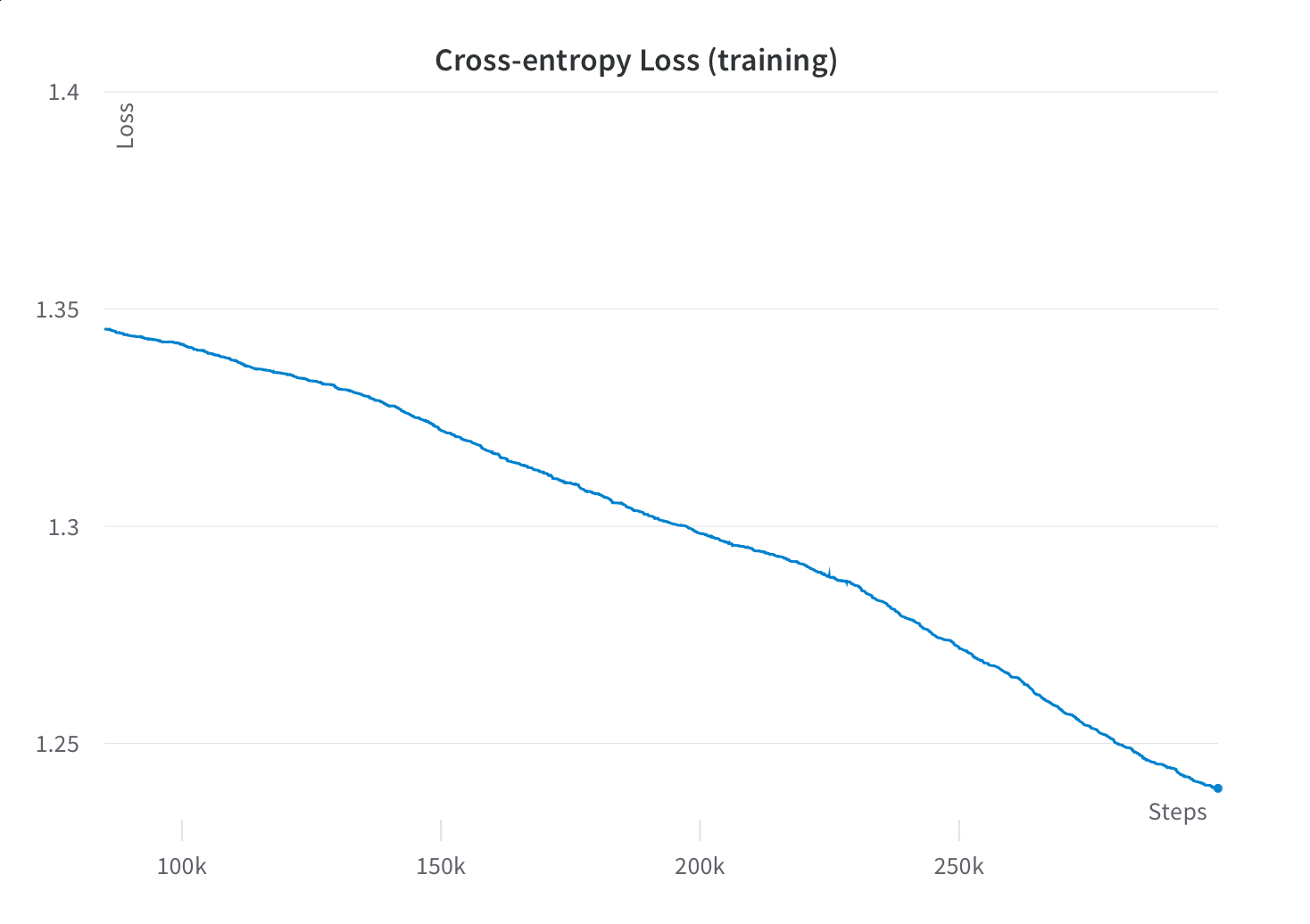
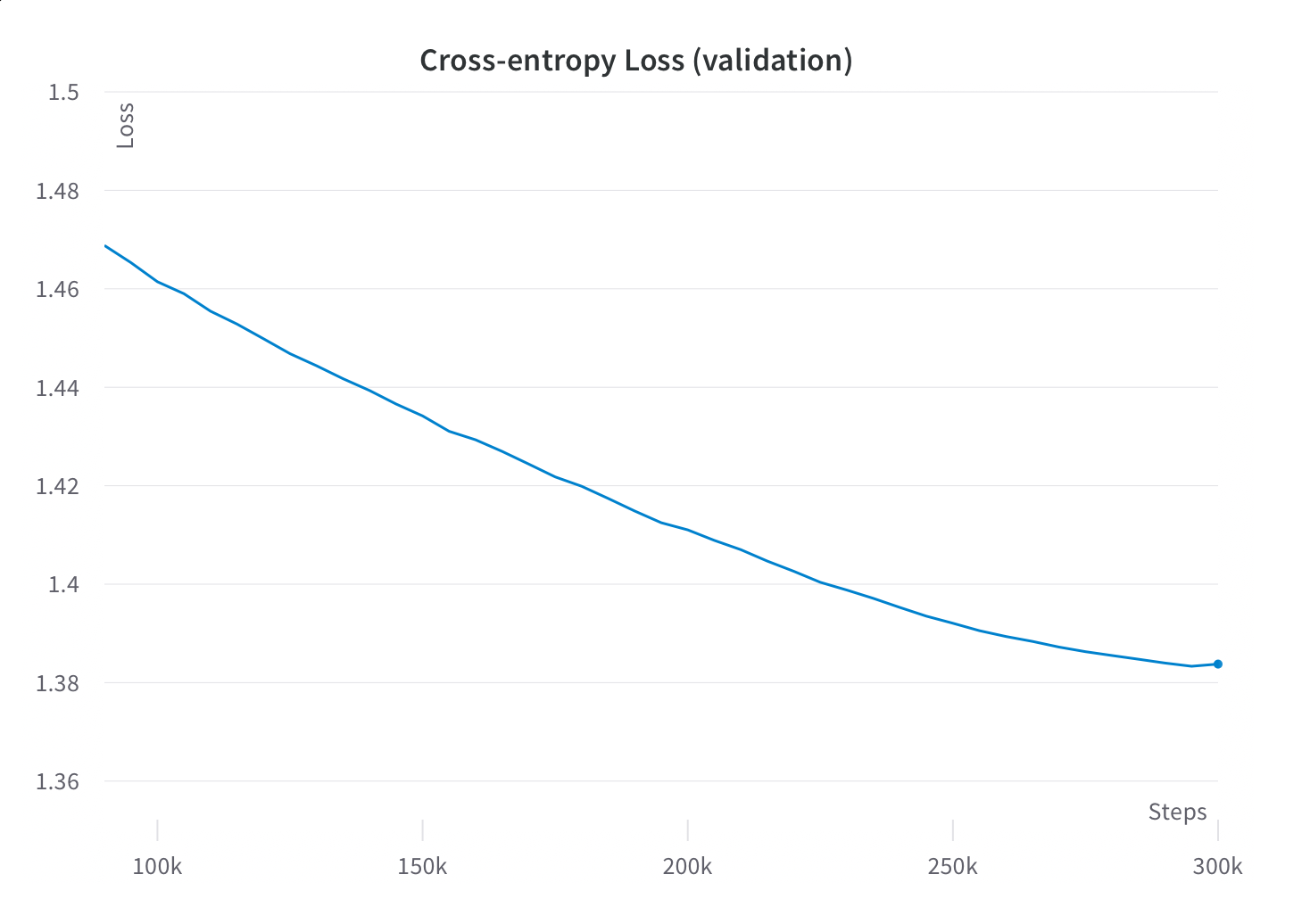
Fortunately, we did continue to see steady perplexity improvements on the validation and training sets for the entirety of training, and preliminary experiments showed improved downstream task performance as we trained out to the full 300B tokens. Our takeaway from this was that it was indeed worth it to train for the full 300B tokens, even though this represented dramatically more passes through the data than comparable models.
Question Answering
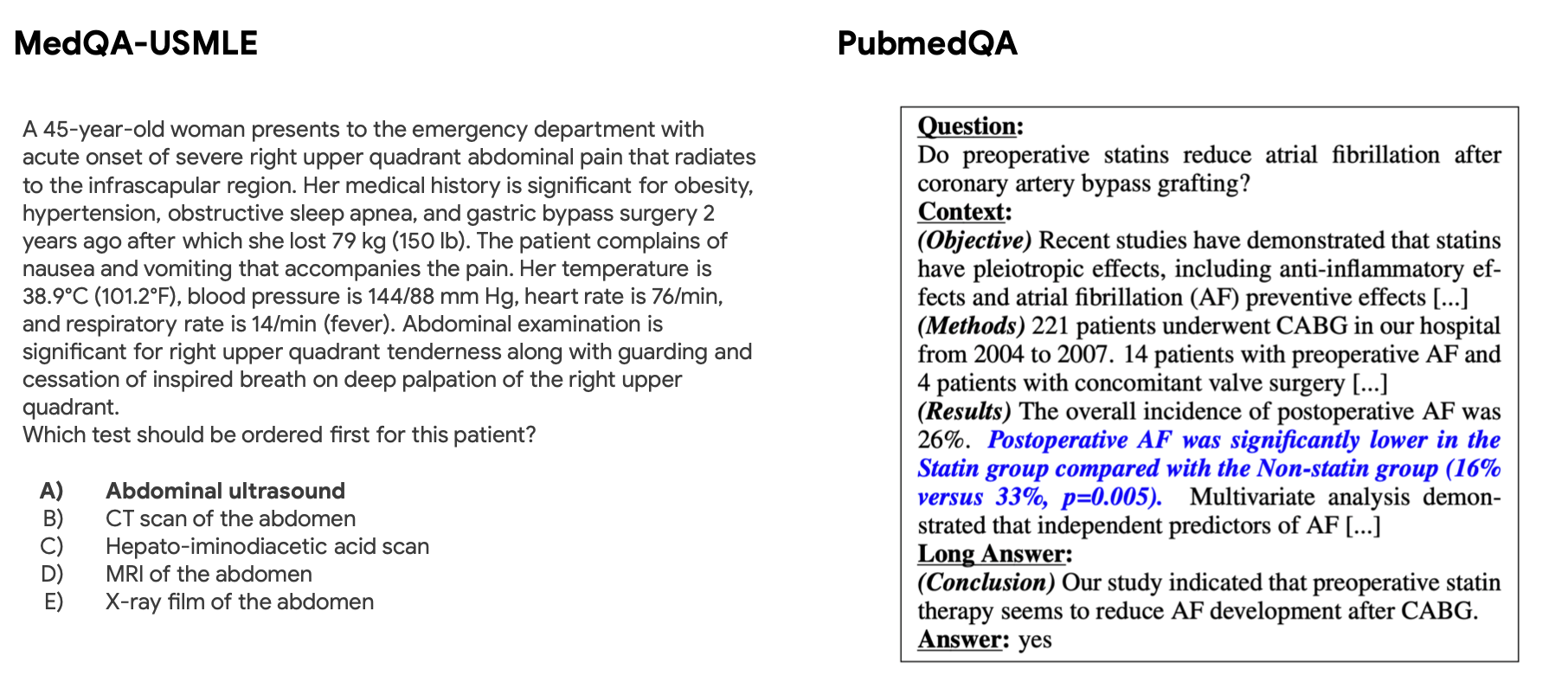
With a fully trained model in hand, we began exploring its capabilities in the biomedical QA space. We focused on 3 common biomedical NLP QA tasks: MedQA, PubMedQA, and BioASQ. MedQA multiple choice questions are derived from online practice exams from the USMLE, the standardized exam used to license doctors in the United States. PubMedQA and BioASQ offer up a passage and ask yes/no/maybe questions based on the passage. The PubMedQA and BioASQ contexts are derived from PubMed abstracts, with questions and answers developed by domain experts.
After full fine-tuning for a small number of epochs, our model is able to achieve competitive results with other state of the art systems, including setting a new state of the art for the MedQA task of 50.3%.
| MedQA | PubMedQA (no answer) | BioASQ | |
|---|---|---|---|
| BioMedLM | 50.3 | 74.4 | 95.7 |
| DRAGON | 47.5 | 73.4 | 96.4 |
| BioLinkBERT | 45.1 | 72.4 | 94.9 |
| Galactica* | 44.4 | 77.6 | 94.3 |
| PubMedBERT | 38.1 | 55.8 | 87.5 |
| GPT Neo 2.7B | 33.3 | 65.2 | 68.3 |
* - zero shot
For each task, our model was fine-tuned with the standard Hugging Face defaults for AdamW using the following hyperparameters:
| Batch size | Epochs | Learning rate | |
|---|---|---|---|
| BioASQ | 8 | 10 | 5e-06 |
| MedQA | 8 | 10 | 2e-06 |
| PubMedQA | 8 | 20 | 2e-06 |
Most of the other competitive systems on these tasks differ from BioMedLM in several ways.
| Model | Scale | Bi-Directional | Extra Data | Lowercased tokenizer |
|---|---|---|---|---|
| BioMedLM | 2.7B | |||
| DRAGON | 360M | yes | yes | yes |
| BioLinkBERT | 340M | yes | yes | yes |
| Galactica | 120B | |||
| PubMedBERT | 300M | yes | yes | |
| GPT Neo 2.7B | 2.7B |
DRAGON and BioLinkBERT are bidirectional systems that leverage additional data (e.g. knowledge graphs, link structure in PubMed). PubMedBERT is a BERT-style model trained on PubMed. Galactica is a GPT-style model trained on scientific literature, while GPT Neo 2.7B is a GPT-style model trained on the Pile (which contains PubMed).
On the one hand BioMedLM has a large advantage in terms of number of parameters versus the smaller bidirectional systems. But there are also other structural disadvantages: it is unidirectional, it does not have access to the richer data used by the other systems, and it does not lowercase the text. In other domains, one may not have access to specialized data like the document link structure and knowledge graphs used by BioLinkBERT and DRAGON. Ultimately these competing systems represent complementary ways to achieve strong performance on domain specific downstream tasks.
Galactica, which is 44x the size of our model and trained on a larger scientific corpus, achieves impressive zero-shot performance on all 3 tasks.
Since BioMedLM is a unidirectional model, it required some extra care during fine tuning. Our experiments showed it was key to use the following format for PubMedQA and BioASQ with the question placed last when presenting the questions to the system:
<Context token> Text of context … <Question token> Text of question <Answer token>
The other bidirectional systems in our table use the PubMed BERT tokenizer, which was trained on PubMed abstracts and lower cases the text. We initially trained a similar GPT tokenizer on the PubMed abstracts and did find that lower casing the text helps on QA performance. We ultimately chose to train a tokenizer that preserves case to balance performance on understanding and generation tasks.
Due to time and resource constraints we did not evaluate other biomedical systems on the above tasks that have not published numbers on those tasks. BioMegatron and GatorTron are large bidirectional models trained by NVIDIA. BioGPT is a smaller unidirectional model similar to ours which is able to score a 78.2 on the version of PubMedQA that includes the answer text in the context.
Text Generation
So far we have explored our model’s generative abilities on several prominent summarization tasks. Particularly intriguing are the system’s performance on the MeQSum task. This task requires the system to read a voicemail/email question from a patient and deliver a succinct summary question. The original questions come from a set provided by the U.S. National Library of Medicine. Three medical experts then manually summarized the questions.
One could imagine an automated question summarization system routing concise questions to doctors or nurses or even another biomedical QA system. (Note: We stress BioMedLM’s generation capabilities are for research purposes and are not ready to be safely deployed in a healthcare setting)
It is unclear that automatic evaluation metrics such as ROUGE properly capture the quality of responses generated by language models for tasks such as this. We believe proper evaluation requires human judgment provided by trained medical professionals, and will be working on conducting such evaluations in the near future. For now we present some examples of our system’s performance on this task:
Example 1
Input
SUBJECT: ball and socket joint MESSAGE: sir, with due respect I would like to state that there is a pain in my ball and socket joint . I didn’t feel and pain in normal or lower position as I up my hard there is a huge site of pain in my ball and socket joint. please prescribe a medicine for the cure .
Gold
What are the treatments for joint pain?
BioMedLM
What are the treatments for ball and socket joint pain?
Example 2
Input
SUBJECT: fluocinolone acetonide ointment tube MESSAGE: Is it possible to obtain this ointment (.25 mg) in anything other than a metal tube? I travel a lot, and the tube constantly develops cracks, so the ointment squeezes out everywhere – very messy. My pharmacy doesn’t know of any other packaging. Thank you!
Gold
Does fluocinolone acetonide ointment.25 mg come in a non-metal tube or other packaging?
BioMedLM
How can I obtain fluocinolone ointment tube (.25 mg) without a metal tube?
Example 3
Input
SUBJECT: Amputed finger MESSAGE: SIr; my 4 yrs old daughter left hand ring finger was crushed in fatal door closure… expert plasti c surgeons were consulted and it was finally decided to remove the dead balck portion of finger and sealed it; now i wish to know to knw wht best treatement I can get for her… she is having pain as her finger/hand is still in plaster…… I need expert opinions ; pl help //// regards [NAME] Father [CONTACT]
Gold
What is the treatment and prognosis for a crushed finger on a child?
BioMedLM
What are the treatments for ring finger amputation?
Example 4
Input
SUBJECT: Coronary Artery Spasms MESSAGE: I ask this as a General Question. Can a Pacemaker help prevent these spasms?
Gold
What are the causes of coronary artery spasms, and can a pacemaker prevent them?
BioMedLM
Can a pacemaker help prevent coronary artery spasms?
Across these varied examples our language model is generally able to produce concise, relevant questions that summarize the patient’s request, not only in the standard case of asking about a treatment for a medical condition, but even handling inputs with detailed real world scenarios.
The model does not always perform well. At times it will create vaguely relevant questions that aren’t correct, leave out key details, and over rely on the pattern “What are the treatments for …” .
Example 5
Input
SUBJECT: Shingles MESSAGE: When can I wash my hair . I have shingles in my head.
Gold
Can I wash my hair if I have shingles on my head?
BioMedLM
What are the treatments for shingles?
Example 6
Input
SUBJECT: family lineage with three types of genetic disorder MESSAGE: In our family line there are three children with different genetic disorders, i want to know if this is a congenital condition that could affect future offspring in this lineage. One has downs syndrome, one had wolf herschhorn, and one has 46xy, 46xxy and 46xxxy; together referred to as Mosaic. they are all third cousins, there are many other children who will one day possibly have offspring is there a danger, should each have genetic testing
Gold
What are the inheritnce patterns of Downs Syndrome, Wolf Hirschhorn Syndrome, and 46xy, 46xxy, 46xxxy mosaicism?
BioMedLM
What are the inheritance patterns of genetic disorders?
This is just the beginning of our investigation of the model’s generative performance. Over the coming weeks we will expand the space of downstream tasks and continue to refine our fine-tuning and evaluation processes for biomedical text generation.
Conclusion
We have just begun to explore the capabilities of this model, and will be continuing to evaluate it on a larger set of biomedical NLP tasks. Meanwhile we invite the community to download the model and start fine-tuning it for your own work. Let us know what tasks you are working on and would like us to look at in the future!
